Introduction
"The underlying
physical laws necessary for the mathematical theory of a large part of physics
and the whole of chemistry
are thus completely known, and the difficulty is only that the exact
application of these laws leads to equations much too complicated to be
soluble." -- P.A.M. Dirac, 1929
Virtually every paper on
computational chemistry begins with this famous quote by the British
theoretical physicist and one of the pioneers of quantum mechanics, Paul Adrien
Maurice Dirac (1902-1984). His
1929 quote that we now “know” chemistry but that the mathematics are not
“soluble” (able to be solved) serves as the foundation for this and every other
paper in computational chemistry.
In this case, to know chemistry means that we can describe it using detailed mathematics
Computational chemistry,
sometimes referred to as molecular modeling or computational quantum chemistry,
represents the newest method of conducting chemical research, joining its
well-established colleagues of observational, experimental, and theoretical
chemistry. Given the increasing prominence of computation in the
chemical profession, this paper looks to provide support for this simple
premise:
An understanding of and ability to use the technologies, techniques, and tools of computational chemistry are as important for chemistry students as those technologies, techniques and tools found in the traditional “wet” laboratory.
A casual scanning of the
scientific literature supports this premise. Computational articles are easy to find in journals such as
the Journal of Computational Chemistry (Wiley Publishers) and the Journal of Computer Aided Chemistry (Japan), but also appear regularly in Science,
Nature, and the Journal of
Chemical Education. Given that reality, it is critically
important that chemical educators at all levels, beginning with pre-college
students, include computation as an essential skill. We as chemical educators would never
ignore the teaching of basic concepts in acid-base chemistry or fail to provide
opportunities for students to learn how to perform a titration or other basic
laboratory skill. Likewise, we can
no longer ignore the prominence of computation as one of the essential tools in
the arsenal of the research chemist.
Computational chemistry must be included in our list of what it means to
know chemistry. Our students must be as familiar with
the Schrödinger equation as they are with the ideal gas law, and must be able
to perform a vibrational analysis of a compound as readily as they are able to
find its experimental melting point.
Likewise, especially in this day of high-stakes testing at the
pre-college level, we must expect the student to demonstrate his or her
proficiency in computational chemistry on end-of-course exams, AP exams, and
other evaluation tools. Without
that expectation, students will not be exposed to these tools and methods, and,
as a result, will not be adequately prepared for careers as scientists.
One might ask: why are
computational methods not a more integral part of the modern chemical curricula
at the undergraduate or pre-college levels? A simple (and perhaps simplistic) reason is offered: teachers teach the way they were
taught. Those of us in our 40s
and 50s are of that generation where computers were not as powerful, nor as
ubiquitous, as they are today. In
my own experience as a computational chemist, the calculations that I can now
perform on a handheld PDA or a low-end laptop were only possible on big
mainframes or supercomputers when I began my career as a computational chemist
in the late 1980s.
Regardless of why computation
is not more prominent, the chemical education profession must begin a serious
effort to increase the importance of computing in chemistry. Organizations such as the National
Science Foundation (NSF) have funded a considerable number of programs at the
undergraduate and pre-college levels to increase an understanding of
computation in all disciplines of science, including chemistry. Programs such as the SuperQuest
Supercomputing Challenge (1987), the National Computational Science Leadership
Program (1998), and other NSF-funded programs have looked to explore what
students are capable of doing and how teachers might be prepared to increase
the role of computing in the science classrooms. NSF’s concern about increasing the number of computationally
savvy science students has even reached into non-traditional audiences. My own funding from NSF has included
significant funds to determine how to incorporate computational science into
classrooms with deaf children by developing the sign language vocabulary needed
to communicate the concepts of computation to that audience.
The remainder of this paper
will provide a short description of computational chemistry, followed by a
description of efforts being made to improve and increase the teaching of
computational chemistry at the pre-college level.
Computational Chemistry Simplified
Simply put, computational
chemistry is the merging of chemistry, computing, and mathematics. In computational chemistry (again, a
term synonymous with molecular modeling), a molecule or reaction mechanism is
studied by applying one or more mathematical theories to determine one or more
properties or behaviors of a molecule or chemical system.
In teaching my students, I
have a number of “sound-bite” phrases and sayings that help them to remember
what they are trying to accomplish, and how they might interpret the
results. The first, and most
important of these, is a daily mantra, and comes from the industrial
statistician George Box: “All
models are wrong, some are useful”.
It is important for students to remember that all descriptions of
chemical systems are simply models, and all make assumptions about those
systems that are not found in reality.
Today’s students have to be warned about accepting answers simply
because the computer says it’s right.
In teaching computational
chemistry as a formal class and in my integration of computational chemistry
into core chemistry classes, I also use the “SPA” framework on an almost daily
basis: Structure-Property-Activity. I teach my students that to really know
a molecule or molecular system, they must understand the structure of the
molecule(s), the property of a molecule (defined as those characteristics of
the molecule that the molecule has by itself), and the activity of a molecule
(those properties that a molecule exhibits in the presence of other
molecules).
I also use a number of analogies to help students understand both the basics of computational chemistry as well as some of the more challenging concepts. I suggest to students that there are five considerations that the computational chemist needs to make in applying computation to the study of a molecule or a molecular system. By way of analogy to cooking chicken, I suggest that the chef needs to know what s/he is trying to prepare, the form of the raw chicken, what ingredients s/he has available, what cooking tools s/he has, and what recipes s/he knows or can find. Analogously, the computational chemist needs to know what s/he is trying to compute, the geometry of the molecule or molecular system, the mathematics available in a given software package, what software is available to that user, and what computational theories the user has and knows. The chart below captures these considerations and their application to both chicken and molecules. Over a span of almost 20 years of teaching computational chemistry to pre-college students, I have found that this analogy provides a great deal of comfort to students when they become overwhelmed with the complexities of a particular computational challenge:

By way of illustration, one of the first computations students perform is a vibrational analysis of water. All molecules vibrate in the infrared, and the number of vibrations can be predicted by various laws such as the “3N-6” rule, where N is the number of atoms (producing, in this case, three vibrations).
In this study, students begin
by building the water molecule and performing a basic optimization of the
geometry to determine the lowest energy value of the molecule. Based on this
optimization, they are able to determine the bond lengths and angles of the
molecule. They then perform a
vibrational analysis of the molecule using the semi-empirical software package
MOPAC1 (Molecular Orbital PACkage), with a PM3 mathematical
parameterization set. Once the
calculation (known as a “job” in computational chemistry parlance) is
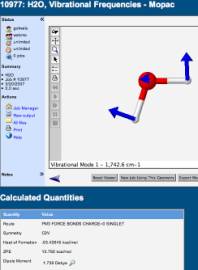
completed, the results are available for analysis. A screen capture of some of the results is shown below. We can see one of the vibrations of
water (scissoring), and some of the properties of the molecule: energy in terms of heat of formation
(kcal/mol), dipole moment (Debyes), and its molecular symmetry (C2V).
It should be noted that the students can see an animation of the vibrations,
not just the static image shown in this screen capture. The students can also rotate the
molecule, zoom in or out, and otherwise see the molecule from a variety of
perspectives. For this particular
calculation (vibrational frequency), students can also view calculated IR
spectra of the water molecule. I
teach them that, depending on the choice of mathematical theory (known in the
community as a model chemistry),
the data from the IR spectra is skewed by about 11% from that of experimental
vibrational frequencies (“all models are wrong....”). They learn how to compare
computed results with that of experimental results by using resources such as
the Computational Chemistry Comparison and Benchmark Database from the National
Institute of Standards and Technology (NIST, http://srdata.nist.gov/cccbdb/). Lively discussions on who is “right”
(experimentalists or “computationalists”) are not uncommon.
Other data generated by this
job includes partial charges and bond orders. Also available to the students is the raw output of the job,
which provides a significant amount of data, much of it beyond the realm of
most pre-college students (and teachers).
On the North Carolina High
School Computational Chemistry Server2 (described in more detail
later in this paper), students build molecules, submit jobs, and analyze
results using an inexpensive Web/Java-based interface known as WebMO3. This interface provides an easy-to-use
connection between the user and the research-grade quantum chemistry software
packages, such as MOPAC, Gaussian 034 and GAMESS5
(General Atomic and Molecular Electronic Structure System).
Readers are strongly invited
to “take a spin” on the server by going to http://chemistry.ncssm.edu,
logging on as guest (password: guest), and, in the vernacular, performing a
Hartree-Fock (HF) 3-21G vibrational analysis of water. Detailed instructions are provided on
the main page in the lab “Case Study:
Vibrational Frequencies of Water”.
The guest account allows users to run very small jobs to get a
sense of what computational chemistry is like, but not run production
(research) level calculations.
Resources for that are described later in this paper.
Assuming that the reader has
a basic sense of what computational chemistry is, we now turn to pedagogical
issues. How does the chemistry
educator use computational chemistry in the classroom? Over the past 20 years, I have taken
three approaches to academic uses of computational chemistry, all at the
pre-college level:
- Integration into the “traditional” classroom
curricula
- Establishment of a stand-alone course in
computational chemistry
- Establishment of a research program in computational
chemistry
Each of these approaches is
described in this paper. Prior to
that discussion, however, a broad-scope picture of pedagogy is perhaps
useful. Computational science educators
call this, with some degree of tongue in cheek, the “associative law” of
computational chemistry education:
Computational (chemistry education)
(Computational chemistry) education
This “law” suggests the
following. There are two ways we
can use computation in chemistry.
The first is as a tool for teaching the same topics that are taught
using more traditional methods:
atomic and molecular structures, acid/base chemistry, kinetics,
thermochemistry, organic chemistry, and the like. Conversely, we also teach students about the technologies, techniques and tools of computation.
What is a semi-empirical method?
How do ab initio methods
differ, both in their mathematics and in the quality of the computation? I believe that learning about computational chemistry methods is as important as
using those methods to learn about
chemistry.
Classroom Integration
My introduction to
computational chemistry came as a result of teaching chemistry to very advanced
students at the Blair Magnet Program at Montgomery Blair High School (Silver
Spring, MD) in the late 1980s. It
was clear that these students had no conceptual idea of what was happening at
the molecular level, especially when addressing topics such as reaction
dynamics, activation energies, and chemical kinetics. Through work with the NSF on a number of high school computational
science programs, we were able to see the power of computing in providing
students with a way to see and study scientific events that happen too fast,
too slow, are too large, or are too small. My experiences with those programs were life changing: I became a full-time computational
science educator, working at a supercomputing center in North Carolina and with
various NSF-supported computational science education organizations.
Now back in the classroom
full-time at the North Carolina School of Science and Mathematics (NCSSM, http://www.ncssm.edu, a state-supported
residential school for high school juniors and seniors with a strong interest
and aptitude for science and math), I integrate computational opportunities on
a regular basis in my AP Chemistry course. The approach I take is described as “just in time”. By this I mean that when the students
are struggling with some concept, I will bring computation in as a tool for
them to better understand that concept.
I do not take time to provide them with a lengthy background overview of
computational chemistry, its varied methods and mathematics, or even its
implementation. I introduce enough information for them to use computational chemistry
as a problem-solving tool. For
example, this coming week (Sept 10, 2007) in AP Chemistry we are beginning a
discussion of aqueous solutions, and I will use a computational analysis of
water to introduce the concepts of molecular structure, bonding, and properties
such as dipole moment and heats of formation. As this is their first exposure to computational chemistry,
I will use the chicken analogy to help them have a sense of what they are doing
and why, but the focus is on using computation to learn all we can about
water. In other words,
computational (chemistry education).
During the course of the
year, I routinely have the students perform computational labs, in addition
to the standard wet lab activities
advocated (mandated?) by the AP Chemistry curriculum. Many of the AP topics are difficult ones for students,
especially those related to atomic and molecular structures. For example, Lewis dot structures and
their significance are conceptually very difficult. With computation, we can mathematically calculate and
visualize lone pair orbitals. A lab (currently in development) looks to help
students understand why some molecules violate the octet rule. Without
computing, students have to take it on faith that not all molecules follow the
rules. In looking at bonding,
students can perform molecular and natural bond orbital calculations and
visualize sigma (s) and pi (p) bonds. In reaction kinetics and thermochemistry, we can
computationally determine the activation energies (Ea) of a
reaction, and determine the rate constant for a particular reaction. We use computation with our organic
chemistry students to help them develop a more intuitive feel for organic
structures and functional groups, and perform computational experiments to
determine the pKa values for carboxylic acids. We teach a course at NCSSM in environmental chemistry, and
students use computation to look at the degradation rates of atmospheric
pollutants. In this activity,
students compare their calculations to the experimentally determined kinetics
of a variety of environmentally relevant compounds.
In the process of integrating
computation into the traditional classroom, we make every effort to use the
right tool for the right job, and work hard to help our students understand why
and how we make those decisions.
Most of my colleagues are traditional experimental chemists, and I am
the only computational chemist on staff.
As such, we work collaboratively to help students decide if a problem is
better solved by running an experiment in the lab, by running a “comp chem
job”, or by some combination of the two.
The experimental chemists, of course, want to address every problem in
the lab, while I tend to run to my computer at the drop of a hat. Because, however, we care about the
best interests of our students, my colleagues will sometimes advocate
computation while I will sometimes advocate a wet lab approach to a chemical
problem!
At NCSSM, all instructors
teach core chemistry courses – honors and AP – but also teach advanced
electives, including organic, analytical, polymer, environmental, and
industrial chemistry. In addition,
we offer a program of instruction in chemical research. NCSSM is on a trimester schedule,
meaning an elective course meets for 10-12 weeks from three to 5.5 hours per week.
Introduction to Computational Chemistry
In addition to the electives
described above, I developed a trimester course “Introduction to Computational
Chemistry”, a 30-hour program of instruction. A partial screenshot of the syllabus is shown below. The reader should note that I use the
same general course description as that of a graduate level course at the
University of Denver.

In this course, which
routinely generates a waiting list of students wishing to participate, the
focus is on both aspects of the “associative law” pedagogical approach. It is my goal in this course to provide
students with the opportunity to learn how computation helps to solve
challenging chemical problems. It
is also my goal to provide students with the opportunity to learn the jargon,
the foundational mathematics, and the methodology of computational
chemistry. While some of our
students will move to the university with the goal of being able to use
computation as a tool for chemical learning and research – computational
(chemistry research) -- all three of our local universities (Duke, University
of North Carolina- Chapel Hill, and North Carolina State University) have
extensive research programs in the development of new methods for doing
computational chemistry. As such,
and since many of our students matriculate to one of those three universities,
it is my goal to prepare them to work in that developmental area of
(computational chemistry) research.
In the Introduction to
Computational Chemistry (Chem 412) course at NCSSM, I ask my students to be
able to provide a meaningful discussion of the following questions by the end
of the course (determined, of course, by the final exam!):
- What is the role and purpose of computational
chemistry? What does
computational chemistry allow us to do that cannot be done using
traditional (i.e. wet) chemistry?
- What is the fundamental mathematical expression
that needs to be solved in doing computational chemistry? What are the terms in this
equation, what is their significance, what variations can be used?
- What are the approximations that can be used in
doing computational chemistry?
What are the pros and cons of the various approximations? How does choice of approximation
affect the results, computing time, etc.?
- There are roughly four different
"flavors" to computational chemistry: ab initio methods, semi-empirical methods, density
functional theory (DFT) and molecular mechanics/molecular dynamics. What
are these methods? How do
they differ?
- What are the fundamental units of measure used by
computational chemists? What
are some different ways that these fundamental units might be expressed?
- What are some of the computer codes that one
might use to do computational chemistry? What platforms are needed for these codes, what are the
strengths and limitations of these codes?
Most if not all of these
questions focus more on the (computational chemistry) education aspect, rather
than the computational (chemistry education) part of the pedagogy. However,
each student, generally working with a team of one or two other students, also
completes a research project on a topic of interest to that group. Most of the students pick topics in
which they are applying some technique of computational chemistry to an
interesting chemical problem, while a few do studies that compare different
computational approaches to some problem.
Students must present their research in journal form, following the
format of the Journal of Computational Chemistry (the same format used in the PDF version of this paper). A “Journal of Student Computational
Chemistry” from the Spring 2007 semester can be found on the Web at http://chemistry.ncssm.edu/JcompChem.pdf.
The students in this class
routinely (approximately once a week) read and discuss an article from the Journal
of Computational Chemistry. Students are expected to learn how to
read the primary scientific
literature in this course, and be able to discuss their readings intelligently
with me and with their fellow students.
The bottom part of the
syllabus showing how this class is structured is provided below. NCSSM is
unusual among high schools in that, since we are residential, we are able to
provide evening classes. My course
is held one night a week for three hours, so I need to structure it such that
students are not subjected to a three-hour lecture! During the course of the class, they have a lecture, do an
in-class lab, break out into small groups to discuss a journal article, and get
instructions for the out-of-class lab that they must complete by the next
week. We start with a 20-minute
quiz on the previous week’s discussions and notes. Students are required to
keep a lab notebook, and learn how a computational lab notebook is similar to
but different from an experimental lab notebook. All NCSSM courses use the course management tool Moodle
(http://moodle.org), an open-source (i.e., free) Web-based tool similar to
Blackboard and WebCT.

Research in Computational Chemistry
Students who wish to continue
their studies in computational chemistry can elect to take Chemistry 414,
Research in Computational Chemistry.
In this course, we explore more advanced techniques and tools, and students
again conduct an independent research project, typically at a higher level than
that of the project done in the Intro course. For this course, with its focus on the software program
Gaussian 03 as the main computational engine, the course uses Exploring
Chemistry with Electronic Structure Methods by James Foresman and Aeleen Frisch as the textbook.

This course has a support
team of computational chemists at both Duke and UNC-Chapel Hill, consisting of
post-doctoral students and research faculty. These researchers provide “just in time” support to me in
answering questions that are out of my area of expertise, or work with students
directly as needed. In addition to
the use of standard computational chemistry tools (Gaussian, GAMESS, MOPAC), I
also introduce other tools, such as AutoDock6, a protein docking
software package developed by the Scripps Institute.
Sample projects for the
current class include one student who working to improve the functionality of
the WebMO interface by writing additional code; a student looking at the molecular
structure of the compound responsible for the generation of oxygen through the
Photosynthesis II process; and a student who has been working experimentally
with the molecule heparin through a research program at UNC-Chapel Hill, and is
doing an computational study of that molecule to augment his experimental
work.
Medicinal Chemistry
seminar
NCSSM also provides students and faculty with the opportunity to offer seminars based on personal interests, usually meeting once a week for 90 minutes. I offer a seminar in Medicinal Chemistry based on work I did earlier in my career in the area of anesthesiology and toxicology. In this seminar, which is completely computationally driven, students learn the basics of pharmacology, spend several weeks in drug design activities, and then spend several weeks looking at pharmacogenomics (using a variety of computational biology tools!). In one of the weekly computational labs, they do a small case study to determine the optimal geometry for the molecule acetylcholine, and conduct a potential energy scan (PES, also known as a coordinate scan) of that molecule:
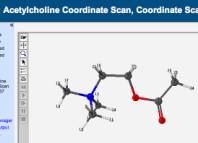
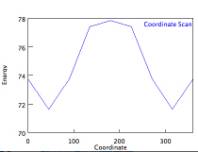
The seminar culminates with a large case study in which students must apply a variety of computational methods to the study of a medicinal compound. Playing one of several roles, students must work as a team to solve a complicated case study. A screenshot of the case study is shown below:

North Carolina’s
Experiment
As mentioned earlier in this
article, the computing tools used by my students were once only available on
high performance computers, and thus off-limits to students, available only to
research university and commercial scientists. As computers became more powerful and software was “ported”
to desktop computers, the possibilities for students increased. At the undergraduate level, more
chemistry departments began to establish computer labs with one or several
computational chemistry software packages.
These tools, however, were
simply too expensive or required computers not available to most high schools,
even specialized schools such as NCSSM.
In my own work, conversations with software vendors typically solicited
the comment that high school students were simply not able to use their software. Even for those vendors (and they were
few) who were supportive of pre-college efforts, they were unwilling or unable
to provide the deep discounts that would be needed for high school
budgets.
A number of efforts, such as
the NSF-supported “ChemViz”7 program at the National Center for Supercomputing
Applications (NCSA), developed access via the Web to computational chemistry
tools and curricula for pre-college students. This resource, highly innovative in its time, did not
provide students with powerful enough tools to do research. Nor did it have sufficient breadth and
depth for teachers to be able to support a program of instruction in
computational chemistry. At NCSSM,
we were able to badger the software vendors enough to get software donated or
at a very low cost. This model,
however, was not sustainable for many high schools.
Over the years, a number of
research-grade codes, such as MOPAC, GAMESS, and others became available for
desktop computers and were in the public domain, meaning no cost. These software programs typically,
however, required a significant understanding of computing systems (typically
Unix/Linux) and also required students to learn a fairly cryptic system to
generate input files for a calculation.
For example, the code below is a sample input file for GAMESS:
$CONTRL SCFTYP=RHF
RUNTYP=HESSIAN
ICHARG=0 MULT=1 COORD=ZMTMPC
$END
$BASIS GBASIS=N21 NGAUSS=3 $END
$DATA
C15H12O6
C1 1
C 0.0000000 0 0.0000000 0
0.0000000 0 0 0 0
O 1.4341000 1 0.0000000 0
0.0000000 0 1 0 0
C 1.3718166 1 114.97905 1
0.0000000 0 2 1 0
C 1.4084832 1 123.81387 1
19.441139 1 3 2 1
C 1.3994155 1 118.83432 1
-179.85275 1 4 3 2
C 1.3841214 1 120.93122 1
-0.9970528 1 5 4 3
C 1.4067731 1 119.36659 1
-0.0927585 1 6 5 4
C 1.3980438 1 121.32708 1
0.8675006 1 7 6 5
H 1.0972631 1 120.91645 1
179.50426 1 8 7 6
O 1.3630382 1 115.97457 1
179.85347 1 7 8 3
H 0.9495488 1 108.20358 1
179.91360 1 10 7 8
H 1.0960879 1 120.74505 1
-179.38483 1 6 7 8
H 1.0973780 1 119.79391 1
179.52810 1 5 6 7
C 1.4733444 1 121.04055 1
176.87024 1 4 5 6
C 1.5473835 1 113.38224 1
-49.011031 1 1 2 3
O 1.4112240 1 110.11977 1
-64.884104 1 15 1 2
H 0.9480935 1 106.05570 1
-173.39745 1 16 15 1
H 1.1178096 1 108.10917 1
175.52041 1 15 1 2
O 1.2169777 1 120.33281 1
148.30413 1 14 15 1
C 1.5065672 1 107.29939 1
-174.25849 1 1 2 3
C 1.3945558 1 118.56470 1
-134.69061 1 20 1 2
C 1.3896935 1 120.57845 1
-179.66709 1 21 20 1
C 1.3987819 1 120.06596 1
-0.3073078 1 22 21 20
C 1.4148287 1 119.48882 1
0.1891373 1 23 22 21
C 1.3950668 1 121.41952 1
45.467224 1 20 1 2
H 1.0984649 1 119.57851 1
0.4094933 1 25 20 1
O 1.3686471 1 122.63812 1
179.88063 1 24 25 20
H 0.9494814 1 107.38800 1
0.5338152 1 27 24 25
O 1.3670389 1 117.63406 1
-179.85748 1 23 24 25
H 0.9495209 1 107.47905 1
-179.52436 1 29 23 24
H 1.0965534 1 120.42401 1
179.99587 1 22 23 24
H 1.0961999 1 120.19317 1
0.4409991 1 21 20 1
H 1.1201156 1 106.49306 1
68.559180 1 1 2 3
$END
Needless to say, having students create these types of input files was, and is, an unrealistic expectation, even for the most advanced students (although we did that for several years, not much fun for all concerned).
So the problems of getting
computational chemistry tools into the hands of students were substantial. Software that was too expensive,
requiring advanced computer knowledge to install, and input files that were
very difficult to create added up to an untenable situation for most
teachers. These problems, however,
were solved (at least for us in North Carolina) with the advent of the WebMO
interface.
WebMO is a Java-based tool
that provides users with a very easy to use interface to a number of
computational packages, including Gaussian 03, MOPAC, GAMESS, and others, such
as Tinker and NWChem. The
interface can be taught in one class session, and provides advanced users with
the option to customize jobs by hand.
Most users, however, use the pull-down menus that come with the software
and that can be customized by the system administrator.
With funding from the
Burroughs Wellcome Fund (http://bwfund.org) and
the North Carolina Science, Mathematics and Technology Center (http://www.ncsmt.org/), we purchased a Dell
dual-processor Linux server, a commercial version of WebMO, and a license for
Gaussian 03, with a total cost of about $5,000. In addition to Gaussian 03, we installed the GAMESS, MOPAC,
and Tinker public domain codes on the server. This resource is now available to any pre-college teacher
and student physically resident in the State of North Carolina. Teachers can request classroom
accounts, where each student receives an amount of computing time, limited to
small jobs (under four minutes) with a total time limit (30 minutes). Students can request research accounts
by submitting a research proposal to me, following the format used to request
computing time on supercomputers and other high-performance (“big iron”)
computing systems.
In the two years since its
establishment, the server has accommodated hundreds of students for more than
16,000 jobs. In addition to
courses offered at NCSSM (and described earlier), classes in computational
chemistry are offered via NCSSM’s extensive distance learning program (http://www.dlt.ncssm.edu/distance_learning/DLWorkshops/ComputationaChemistry_Fall-07.pdf).
The main advantage of this
system is that all of the installation and maintenance of the system is done by
NCSSM scientists and system administrators, and is provided to all schools at
no cost. In addition, schools do
not have to install any software, often a problem in North Carolina’s public
schools. Schools only need access
to Java-enabled Web browsers, a technology available to the majority of North
Carolina’s schools.
In working to get more
teachers aware of and able to use the resources, I offered (and continue to
offer) workshops at various conferences such as the North Carolina Science
Teachers Association annual conference.
In talking with teachers, it was clear that there was limited travel
time and funding to attend workshops to learn how to do computational
chemistry. To attempt to address
that issue, and with additional generous funding from BWF and the NCSMT Center,
a colleague (Dr. Shawn Sendlinger at North Carolina Central University) and I
collaborated to write an electronic textbook entitled A Chemistry Educator’s
Guide to Molecular Modeling (http://chemistry.ncssm.edu/book). This book (with some chapters still in
writing) looks to provide teachers with enough background in both areas of
computational chemistry (methodology and pedagogy) to integrate computation
into their own classrooms. I use this
book in my “Introduction to Computational Chemistry” course, in spite of the
fact that it is written primarily for chemistry educators.
National Resources: The Global Grid Exchange
In addition to doing
workshops and distance learning programs across the state of North Carolina, I
have been giving talks and workshops outside of North Carolina. During these workshops, the main focus
has been on how participants might set up a WebMO-based server for their own
schools, region, and/or state.
After several of these workshops, I was able to arrange for seed funding
from BWF to help others who might wish to replicate the North Carolina
resources.
About the same time, Parabon
Computation, Inc. (http://www.parabon.com/), a grid computing organization
based in Reston, VA, contacted me about providing grid resources for
educational purposes. In grid
computing, computing jobs are distributed to idle computers, to be run when
those personal computers are not in use by the owner. The results of the job are then sent back to the host
computer for analysis by the originator.
With support funding from
Cisco Systems, we established a mirror site (http://cli.globalgridexchange.com/)
of the North Carolina machine. On
the grid computer, we have installed the WebMO interface and access to GAMESS,
MOPAC, and Tinker, along with a link to the electronic textbook, labs, and
other resources. Due to licensing
constraints, Gaussian 03 is not available on this machine.
This new and greatly
appreciated resource now allows NCSSM to provide free classroom and student
research accounts to any pre-college teacher or student in the United States.
Professional staff at NCSSM and Parabon maintain both the North Carolina and
national servers, with additional support from a team of six computational
chemistry high school interns at NCSSM.
Even with these resources in
place, it is clear that many teachers will want and need additional support
prior to implementing computational chemistry in their classrooms. We are hoping to be able to provide
significant support, in the form of FAQs (frequently asked questions) pages, an
increasing number of ready-to-go labs, and the sharing of the curricular
materials developed over the past many years. With these resources, I am hopeful that teachers will feel
empowered to add a few activities a year to their classes, and perhaps consider
offering a course in computation at their schools. I am also hopeful that newly trained teachers will develop
their own computational chemistry classroom and lab activities to be shared
with the larger community.
Conclusions
The main premise of this paper is that it is critically important that tomorrow’s chemical scientists understand the technologies, techniques and tools of computational chemistry. With the resources now available – access to research-grade hardware and software at no cost, an ever-increasing body of labs and curricular materials, and other support mechanisms – it is the hope of this author that computational chemistry will as familiar to tomorrow’s chemistry researchers and teachers as the test tube and the beaker are now.
Acknowledgements
Appreciation is extended to
the Burroughs Wellcome Fund and the North Carolina Science, Mathematics and
Technology Center for their funding support for the North Carolina High School
Computational Server.
Appreciation is also expressed to the Global Grid Exchange, Parabon
Computation, Inc. (http://www.parabon.com/)
and Cisco Systems (http://www.cisco.com/) for their support of the national
computational chemistry server.
References
1. MOPAC Version 7.00, J. J. P. Stewart, Fujitsu Limited, Tokyo, Japan.
2. The North Carolina High School
Computational Chemistry Server, http://chemistry.ncssm.edu.
3. Schmidt, J.R.; Polik, W.F. WebMO Pro, version 7.0; WebMO LLC: Holland, MI, USA,
2007; available from http://www.webmo.net (accessed April 2007).
4. Gaussian 03, Revision C.02,
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R.
Cheeseman, J. A. Montgomery, Jr., T. Vreven, K. N. Kudin, J. C. Burant, J. M.
Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G.
Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota,
R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai,
M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, V. Bakken, C. Adamo,
J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi,
C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador,
J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain,
O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz,
Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A.
Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A.
Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B.
Johnson, W. Chen, M. W. Wong, C. Gonzalez, and J. A. Pople, Gaussian, Inc.,
Wallingford CT, 2004.
5. "General atomic and molecular electronic structure
system, M.W.Schmidt, K.K.Baldridge, J.A.Boatz, S.T.Elbert, M.S.Gordon,
J.H.Jensen, S.Koseki, N.Matsunaga, K.A.Nguyen, S.J.Su, T.L.Windus, M.Dupuis,
J.A.Montgomery, Journal of Computational Chemistry, vol. 14, pages 1347-1363,
1993."
6. Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R,
Hart, W.E., Belew, R.K., Olson, A.J. (1998) Automated docking using Lamarckian
genetic algorithm and an empirical binding free energy function. J. Comp. Chem.
19:1639-1662.
7. ChemViz, National Center for Supercomputing
Applications (NCSA), University of Illinois at Urbana-Champaign,
http://chemviz.ncsa.uiuc.edu/