Learning outcomes
The goal of this activity is twofold:
- Computational (chemistry education): Use interactive computing (specifically computational chemistry) to introduce and/or reinforce basic chemical principles including point VSEPR geometry and point group symmetry.
- (Computational chemistry) education: Help students understand how COMPUTATION is used in chemistry education and chemistry research. In this case, students use a high-performance computational chemistry server, running state-of-the-art chemistry software. This server is located in Durham, NC, and is accessible 24/7 to pre-college students who reside in the State of North Carolina.
Teacher planning
Time required for lesson
The computing time for this lab is determined by the specific molecules chosen by the instructor. Most relatively simple molecules, such as PCl5, do not require significant amounts of time to run. Molecules that are larger (20 atoms and up) will require more compute time. It is recommended that students build these molecules, submit the jobs, and obtain their data on the following day.Materials/resources
Access to Java-enabled web browser. Internet Explorer on Windows-based machines is often problematic. Users are encouraged to use Mozilla Firefox, which is downloadable free of charge.
If this activity is used as a demo, the teacher needs an account on the North Carolina High School Computational Chemistry Server. Accounts are free of charge, and instructions for obtaining an account are available at this site. An LCD projector connected to an internet-accessible computer is required for demo purposes.
If this activity is a student lab, students must have accounts on the chemistry server. Accounts are free of charge, and instructions for obtaining accounts is available on the North Carolina High School Computational Chemistry Server.
Technology resources
As described in the Materials Needed section.
Pre-activities
Students should be able to draw and understand Lewis dot structures, and determine the VSEPR geometry of a given molecule.
Activity background:
In chemistry, knowing the structure of a molecule is exceptionally important. To be able to describe the structure of a molecule in language that others understand means that you are “fluent” in chemistry. Knowing the “jargon” of chemistry is one of the goals of any quality class in chemistry, regardless of the level of that class.
Most chemistry classes teach a system of notation for molecular structures known as “VSEPR” -- Valence Shell Electron Pair Repulsion. This system states that electrons are located in specific locations in the molecule, and these electron groupings repel each other. Electrons reside in valence shells in pairs, and these pairs repel each other. Hence, the name VSEPR. The location of these pairs forces the individual atoms in the molecule to move in such a way as to minimize the interaction of one electron pair with another.
The graphic below shows the various geometries. In the chart, the letter “X” represents an atom attached to a central atom designated with an “E”. Lone pairs – those electron pairs that do not form a bond – are represented by two black dots. The triangle wedge shapes between atoms are used to represent the geometry of the molecule. Wedges that are a series of lines suggest that the atom is going away from you, whereas the plain wedge states that the atom is coming towards you. The steric number represents the number of attachments to the central atom. These attachments can be another atom or one or more lone pairs. Based on the steric number and the number of lone pairs, one can specify the name of the molecular geometry. We also use the “AXE” notational system, where “A” is the central atom, “X” are atoms attached to the central atom, and “E” is the number of lone pairs. For example, the bent molecule with a steric number of 4 and two lone pairs would be represented as “AX2E2”. As a chemist, you could communicate this geometry by saying it has “AX2E2” geometry, or, more simply, by saying that it has a bent geometry.
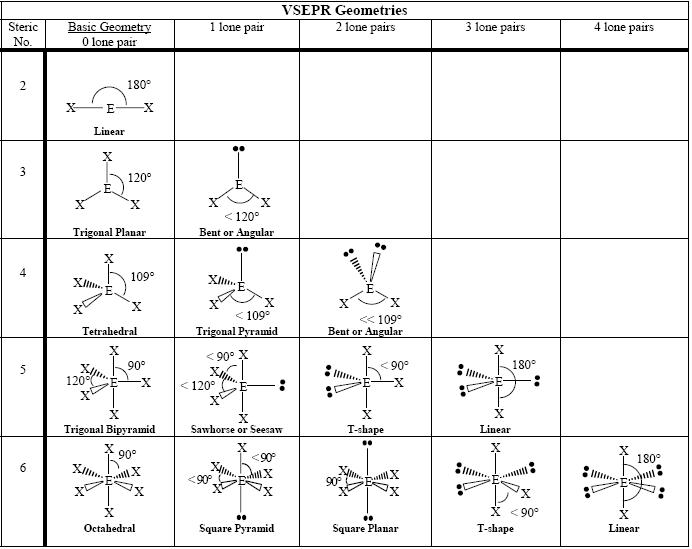
This notational system works very well as a communication tool to describe the geometry of a molecule. To determine the number of lone pairs, one draws a Lewis dot structure for the molecule. This lab assumes that the student can draw these structures. Drawing Lewis dot structures is, however, a relatively tedious endeavor, and we would like to be able to determine the molecular geometry of a molecule without determining its Lewis structure.
Computational chemistry can help with this challenge. All of the modern computational chemistry software packages – Gaussian '03, MOPAC, GAMESS, etc. -- routinely generate what is known as a “point group symmetry” determination for every molecule that is evaluated. Point group symmetries are closely analogous to the VSEPR geometries, but use a different, and more complicated, notational system. The various notational terms include items such as C2v, D3h, and Td. These various terms are determined by a careful analysis of the various ways that the atoms in a molecule can be rotated, reflected through a plane of symmetry, and otherwise manipulated. Being able to determine a point group symmetry by hand is exceedingly more complicated than determining the AXE notation for a given molecule. Point group symmetries are, however, more commonly found in the chemical literature than are VSEPR notations. There are a number of excellent tutorials, such as http://www.reciprocalnet.org/edumodules/symmetry/index.html.
Assuming, however, that we don’t want to learn how to determine point group symmetries by hand, and we don’t want to draw Lewis structures for every molecule we study, how can computational chemistry help us? As before, all computational chemistry software packages determine the point group symmetry of a molecule. For most scientists working in chemical research, knowing the point group symmetry of a molecule is more than adequate for communicating with other scientists. For example, the graphic below shows the molecule PCl5, phosphorus pentachloride. The VSEPR notation for this molecule is AX5, meaning it has five atoms (Cl) attached to the central atom (P), with no lone pairs, as determined by a Lewis dot structure analysis. Based on this determination, the VSEPR geometry of this molecule is trigonal bipyramidal. As you can see under the label “symmetry”, this molecule has a D3h symmetry. This means that the molecule belongs to a geometric family of “dihedral” molecules with three rotational modes through a horizontal plane (i.e., 3h). The various tutorials illustrate these rotations well, and the curious reader is strongly encouraged to go through these tutorials.
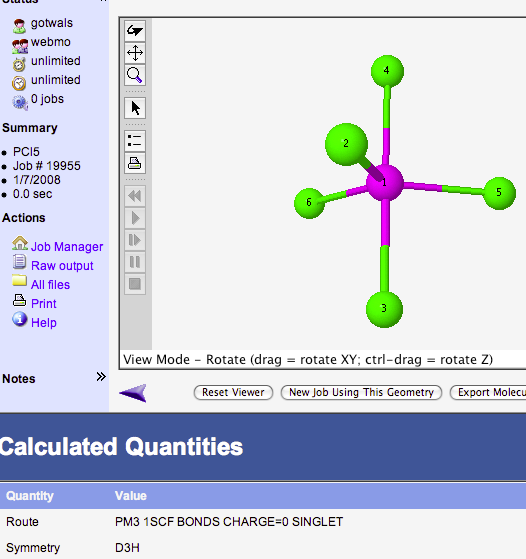
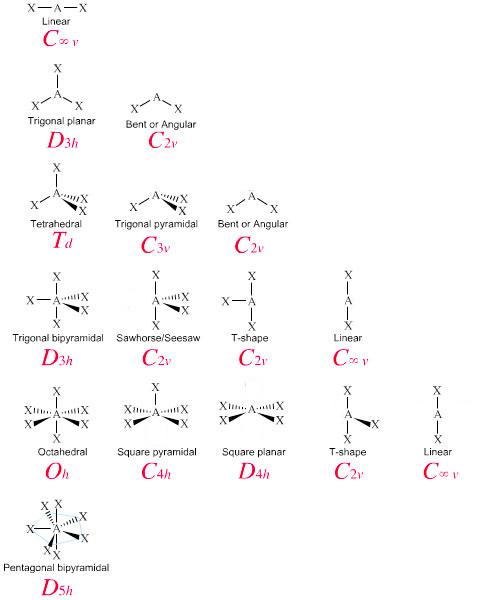
The graphic below shows the correlation chart between point group symmetry notations and VSEPR notations. For example, since the PCl5 molecule has been determined to have D3h symmetry, the chart tells us that it has trigonal bipyramidal geometry using VSEPR and Lewis dot structure notation. Using AXE notation, we identify this molecule has being AX5, with five atoms attached to the central atom with no lone pairs. From the VSEPR chart, this also corresponds to trigonal bipyramidal.
Based on this reading, you should now be able to implement the computational approach procedure described below and determine both the point group symmetry and the VSEPR geometry of any molecule.
TECHNICAL NOTE: the computational chemistry software packages represent the infinity (“∞”) symbol in the point group symmetry chart with an asterisk (“*”).
TECHNICAL NOTE: linear molecules in VSEPR theory can also be correlated to D∞h (D*h) point group symmetry
Activities
In this lab, students will build several molecules, either the ones given in the table or ones assigned by the teacher, and run a basic computational chemistry calculation (using the Hartree-Fock method with 3-21G mathematics with the Gamess or Gaussian software, or PM3 for MOPAC) to calculate the point group symmetry and the VSEPR geometry of the molecule. The specific calculation is known as a "Geometry Optimization".
Method
The following procedure is identical for all molecules. For demonstration purposes, this lab uses the molecule phosphorus pentachloride (PCl5).
Building the Molecule
- Log into the North Carolina High School Computational Chemistry Server.
- In the Job Manager, click on “New Job”.
- Once the molecular editor opens, click on “Open Editor” at the bottom of the window. The Java label should appear. On most machines, it will take Java a few minutes to open.
- A drawing window should open. Click on “Other”, and a periodic table window should appear. Alternatively, you can click on the periodic table icon on the left menu bar. Choose “P” for phosphorus.
- Click once in the drawing window. A purple phosphorus atom should appear. If more than one atom appears, select“File→New”, then select “Tools→Build”, then try again.
- Open the periodic table again, and choose Cl (chlorine). Place 5 chlorine atoms around the phosphorus atom. Connect each chlorine to the phosphorus by drawing a line between each pair of atoms (click-hold-drag).
- Under “Cleanup”, perform a comprehensive cleanup. This should make your molecule look more symmetrical.
- Close the builder window by choosing “File→Close”. Alternatively, you can simply click back into the main window. Make sure your molecule shows up before proceeding!
- To move to the next step, click on the “next” arrow at the bottom right of the window.
Configuring the Job
To prepare your molecule for calculation, follow these steps:
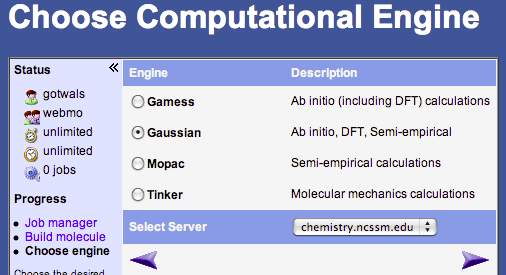
- Choose the computational engine: → any will do except Tinker. MOPAC typically runs the fastest. Once you have chosen an engine, click on the bottom right arrow to go to the next window.
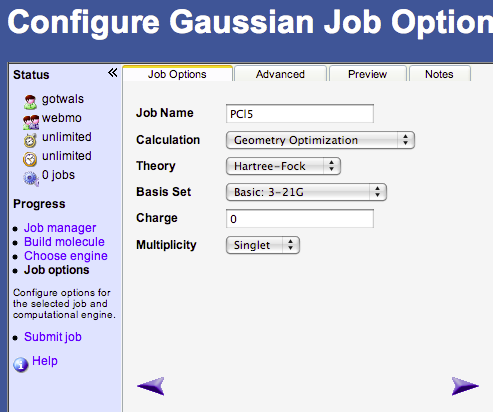
- In the Job Option window you will need to choose the following options:
- Job Name → whatever you want to call your calculation
- Calculation → Drop down menu, choose Geometry Optimization
- Theory → Choose Hartree-Fock for GAMESS or Gaussian, PM3 for MOPAC
- Basis Set → Basic: 3-21G for GAMESS or Gaussian
- Charge → 0
- Multiplicity → Singlet
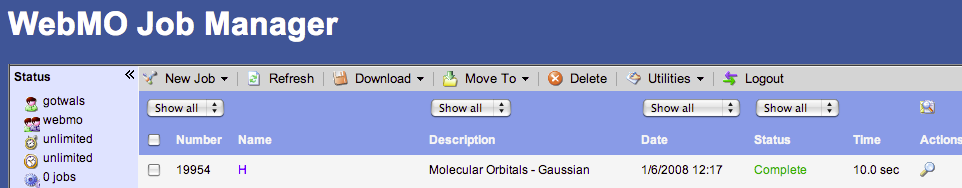
- Click the “next” arrow in bottom left hand corner; this should take you back to the job manger window. You should see your job running in the window or queued up.
- You can click on the word “Refresh” to see the progress of your calculation. This is particularly helpful if you are in the “queue”.
- Once your job is completed, click on the name of the job, scroll down to find the point group symmetry for your molecule (labeled as “Symmetry”).
Analysis of Results
Once your “job” is completed, follow this procedure:
- To view the results of your completed job you may click on the name or the magnifying glass at the end of the job.
- A downloadable student datasheet includes a number of interesting molecules for you to try. You should be able to complete this chart. Alternatively, you can perform molecular geometry determinations of molecules assigned to you by your instructor.
TECHNICAL NOTE: the computational chemistry software packages represent the infinity (“∞”) symbol in the point group symmetry chart with an asterisk (“*”).
Extension
Assessment
Requests for other labs should be sent to Bob Gotwals.
Associated Files
downloadable student datasheet
Modifications
Students are welcome to try their hand at building other molecules and performing similar computations to the one described in this lab.
Critical vocabulary
Important definitions are available via Wikipedia and are linked to this lesson plan.
- computational chemistry
- VSEPR theory
- valence shell
- steric number
- lone pair
- Lewis dot structure
- molecular symmetry (point group symmetry)
- Gamess software
- Gaussian software
- MOPAC software
Comments
It is the belief of all of the major scientific organizations, including the National Science Foundation, that students should develop and demonstrate competence and competence in the following areas:
- Observational science
- Experimentational science
- Theoretical science
- Computational science
This activity makes use of observation, theory, and computation. Students can, on their own, build and calculate other molecules at school or at home. Students can also request a research account on the NC High School Computational Chemistry server, for use in science fair or other research programs.
North Carolina Curriculum Alignment
Science
Grades 9–12 — Chemistry
- Goal 2: The learner will build an understanding of the structure and properties of matter.
- Objective 2.07: Assess covalent bonding in molecular compounds as related to molecular geometry and chemical and physical properties.
- Molecular.
- Macromolecular.
- Hydrogen bonding and other intermolecular forces (dipole/dipole interaction, dispersion).
- VSEPR theory.
- Objective 2.07: Assess covalent bonding in molecular compounds as related to molecular geometry and chemical and physical properties.


